Multiple Sequence Alignment
1. What is a Multiple Sequence Alignment?
Multiple sequence alignment (MSA) is the process of aligning three or more biological sequences (protein or nucleic acid) to identify regions of similarity that may indicate functional, structural, or evolutionary relationships.
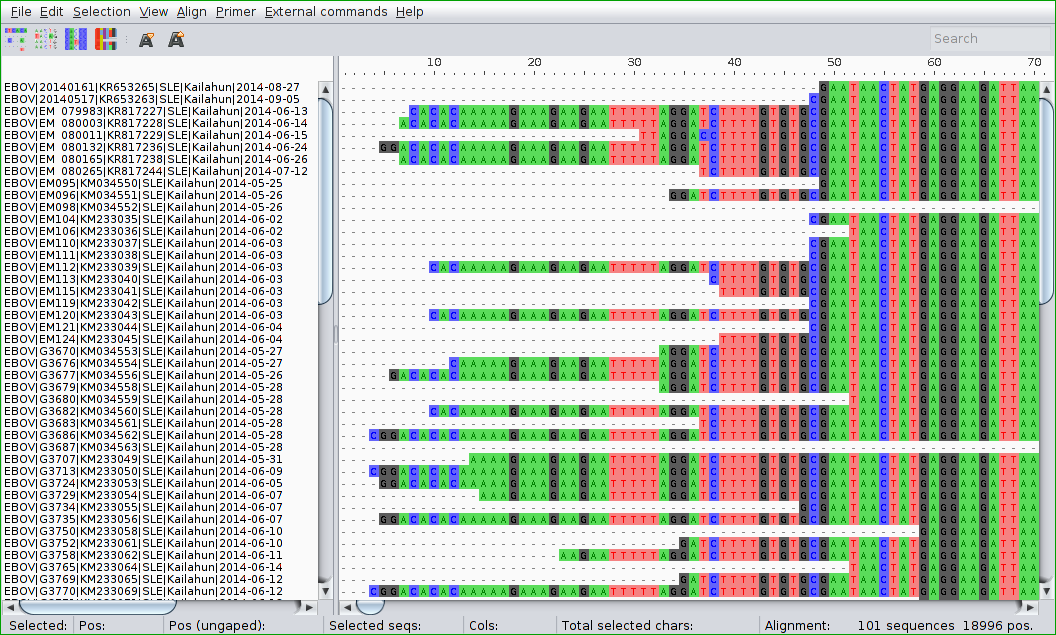
Given sequences $X^{(1)},\ldots,X^{(N)}$ of lengths $n_1,\ldots,n_N$, we seek aligned sequences $A^{(1)},\ldots,A^{(N)}$ of length $n\geq\max\{n_i\}$ such that:
- We can obtain $X^{(i)}$ from $A^{(i)}$ by removing gap characters
- No columns contain all gaps
- The score of the alignment is optimal
Notation
Throughout this lecture, we'll use the following notation:
- Sequence $i$:
- $X^{(i)} = (x^{(i)}_1,x^{(i)}_2,\ldots,x^{(i)}_{n_i})$
- Row $i$ in alignment:
- $A^{(i)} = (a^{(i)}_1,a^{(i)}_2,\ldots,a^{(i)}_n)$
- Column $j$ in alignment:
- $A_j = (a_j^{(1)}, a_j^{(2)}, \ldots, a_j^{(N)})$
Why Multiple Sequence Alignment?
Multiple sequence alignments are essential for:
- Identifying conserved regions across species
- Predicting protein structure and function
- Constructing phylogenetic trees
- Identifying functional motifs and domains
- Designing PCR primers
- Understanding evolutionary relationships
2. Scoring Multiple Sequence Alignments
Sum of Pairs (SP) Scoring
The most commonly used scoring function for MSAs is the sum of pairs method, which calculates the total score as the sum of all pairwise alignment scores.
For column $i$ in the alignment:
where $s(a,b)$ is the pairwise substitution score between characters $a$ and $b$.
Example
Consider the following alignment:
A-CTCAT A-GTC-T ACGTC-T
Let's calculate scores for specific columns:
- Column 3 score: We have C, G, G
- $s(C,G) + s(C,G) + s(G,G) = \text{mismatch} + \text{mismatch} + \text{match}$
- Total = $2 \times \text{mismatch score} + \text{match score}$
- Column 6 score: We have A, -, -
- $s(A,-) + s(A,-) + s(-,-) = 2 \times \text{gap penalty} + s(-,-)$
- Note: $s(-,-)$ is typically 0 (no penalty for aligning two gaps)
Problems with Sum of Pairs Scoring
- Substitution scores were derived as log-odds scores for pairwise comparisons
- The mathematically correct approach would use log-odds scores for triples, quadruples, etc.
- No probabilistic justification for the sum of pairs approach
Mathematically, the issue is that:
Tree-based Scoring
A more biologically motivated approach considers the evolutionary tree relating the sequences:
Tree-based scores are thought to be more biologically appropriate, but they have practical limitations:
- We don't know the true evolutionary tree
- We need to infer ancestral characters at internal nodes
- Different parts of the alignment may have different evolutionary histories (due to recombination)
Despite its theoretical limitations, sum of pairs scoring is almost always used in practice due to its computational simplicity.
3. Multidimensional Dynamic Programming
The Naïve Approach
We can extend the pairwise alignment dynamic programming approach to multiple sequences. Define $F(i_1,i_2,\ldots,i_N)$ as the score of the best alignment up to positions $i_1, i_2, \ldots, i_N$ in sequences $1, 2, \ldots, N$.
The recurrence relation becomes:
Computational Complexity
The naïve dynamic programming approach has prohibitive computational requirements:
- Space complexity: $O(n^N)$ - need to store $\prod_{q=1}^{N}n_q$ values
- Time complexity: $O(2^N n^N)$ - each cell requires maximizing over $2^N-1$ possibilities
Example Calculation
For an alignment of 5 sequences, each 100 characters long:
- Space needed: $101^5 \approx 10^{10}$ numbers
- With 32-bit integers: ~39 GB of memory
- Time: Must compute $10^{10}$ cells, each requiring comparison of $2^5-1 = 31$ values
The MSA Algorithm
Carrillo & Lipman (1988) developed an improved algorithm that uses bounds on pairwise alignments to reduce the search space:
While this is a significant improvement, it's still impractical for most real-world alignment problems with more than a few sequences.
4. Progressive Alignment Methods
Given the computational limitations of exact algorithms, practical MSA methods use heuristic approaches. The most successful of these is progressive alignment.
General Strategy
Key decisions in progressive alignment:
- Order of alignments: Which sequences to align first?
- Alignment strategy: Allow only sequence-to-group, or also group-to-group?
- Scoring method: How to score alignments involving groups?
Guide Trees
Progressive alignment algorithms use "guide trees" to determine the order of alignment:
UPGMA Tree Construction
The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) is commonly used to build guide trees:
UPGMA Example
Given the distance matrix:
| A | B | C | D | |
|---|---|---|---|---|
| A | - | 4 | 8 | 8 |
| B | - | 8 | 8 | |
| C | - | 6 | ||
| D | - |
Algorithm steps:
- Find minimum distance: d(A,B) = 4
- Join A and B to form cluster E
- Recalculate distances to E
- Repeat until all sequences are joined
The Feng-Doolittle Algorithm
Published in 1987, this was one of the first practical progressive alignment algorithms:
- Calculate pairwise distances: Align all pairs of sequences and convert alignment scores to evolutionary distances:
$$d(k,l) = -\log\frac{S(A^{(k)},A^{(l)})-S_{\text{rand}}}{S_{\text{max}}-S_{\text{rand}}}$$
- Build guide tree: Use UPGMA or similar clustering algorithm
- Progressive alignment: Starting from the leaves, align sequences following the guide tree
Sequence-to-Group Alignment
Group-to-Group Alignment
Progressive Misalignment
A major limitation of progressive alignment is that errors made early in the process cannot be corrected later:
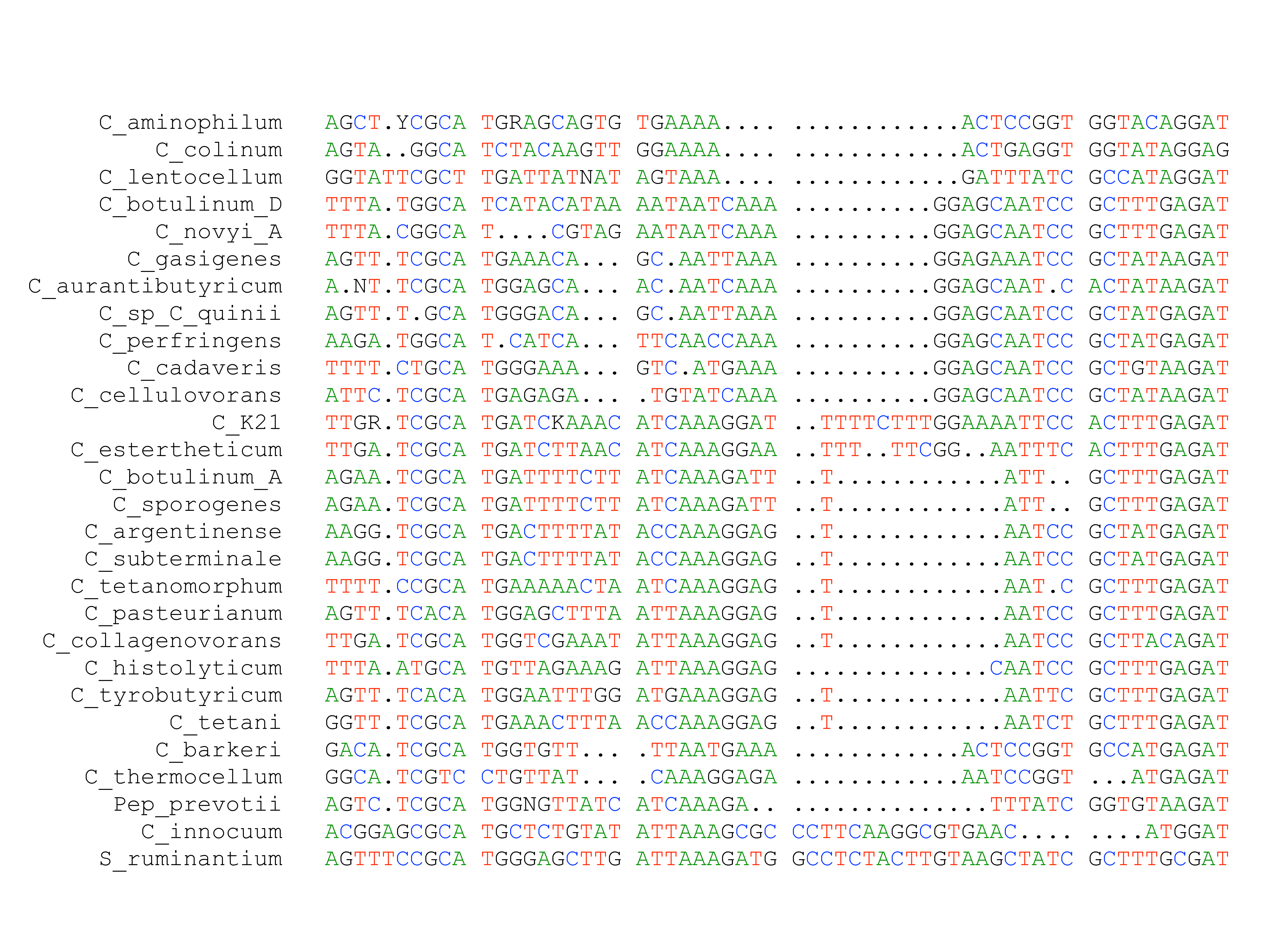
Profile Alignment
Modern progressive alignment methods use profile alignment to align groups of sequences:
The total alignment score becomes: $S(A) + S(B) + S(A \times B)$
CLUSTALW
Thompson, Higgins, and Gibson (1994) developed CLUSTALW, which became one of the most widely used MSA programs (with over 55,000 citations!).
CLUSTALW is essentially the Feng-Doolittle algorithm with profile alignment and many additional heuristics:
- Calculate all pairwise distances
- Construct guide tree using UPGMA or neighbor-joining
- Progressively align using sequence-sequence, sequence-profile, and profile-profile alignment
Modern Progressive Alignment Tools
- Clustal Omega: Successor to CLUSTALW, faster and more accurate
- MUSCLE: Uses iterative refinement after initial progressive alignment
- MAFFT: Offers various algorithms optimized for different scenarios
5. Iterative Refinement Methods
Iterative refinement methods attempt to improve an initial alignment by making small changes and accepting improvements (hill climbing to a local optimum).
The Barton-Sternberg Algorithm (1987)
- Find the two most similar sequences and align them
- Find the sequence most similar to the profile of the current alignment and add it
- Repeat step 2 until all sequences are included
- Refinement phase: Remove each sequence and realign it to the profile of the others
- Repeat step 4 until convergence or a fixed number of iterations
Other Iterative Approaches
- Simulated annealing: Accept some score-decreasing moves to escape local optima
- Genetic algorithms: Maintain a population of alignments and evolve them
- Hidden Markov Models: Use probabilistic models for alignment
6. Practical Considerations and Limitations
General Limitations
- These algorithms maximize match scores, but the "best" scoring alignment may not be biologically correct
- Progressive alignments deteriorate as more sequences are added
- Early mistakes in progressive alignment are frozen and cannot be corrected
- Manual correction is often necessary for alignments of divergent sequences
Statistical Approaches: Fitting vs. Modeling
There are two philosophical approaches to sequence alignment:
Statistical Fitting
- Count change frequencies in real data
- Build empirical descriptions (e.g., BLOSUM62)
- Use log-odds ratios for scoring
- Apply in ad hoc algorithms (BLAST, ClustalW)
Probabilistic Modeling
- Define evolutionary process models
- Specify substitution and indel rates
- Estimate parameters using likelihood/Bayesian methods
- Co-estimate alignment and phylogeny
BAli-Phy: A Bayesian Approach
Suchard and Redelings (2006) developed BAli-Phy, which jointly estimates alignment and phylogeny using Bayesian inference:
This approach is philosophically optimal but computationally intensive, limiting its use to smaller datasets.
Summary
- Scoring: Multiple sequence alignments are typically scored using sum of pairs, despite theoretical limitations
- Exact algorithms: Dynamic programming can find optimal alignments but scales as $O(n^N)$, making it impractical for most problems
- Progressive alignment: Practical heuristic that builds alignments incrementally using a guide tree
- Iterative refinement: Can improve initial alignments but may still converge to local optima
- Trade-offs: Current methods balance biological accuracy with computational feasibility
- Future directions: Bayesian approaches offer principled solutions but need computational improvements
- Durbin et al. (1998) "Biological Sequence Analysis" - Chapter 6
- Thompson et al. (1994) "CLUSTALW" - Nature Protocols
- Notredame (2007) "Recent evolutions of multiple sequence alignment algorithms" - PLOS Comp Bio
Check Your Understanding
- Why is sum of pairs scoring theoretically problematic?
- What makes exact dynamic programming impractical for multiple sequences?
- How do guide trees influence progressive alignment quality?
- What is the "once a gap, always a gap" rule and why is it used?
- When might you choose iterative refinement over simple progressive alignment?