The Coalescent
1. Introduction to the Coalescent
The coalescent is one of the most important theoretical frameworks in population genetics and molecular evolution. It provides a mathematical description of how genetic lineages merge backward in time.
Data: a small genetic sample from a large background population
The coalescent:
- Is a model of the ancestral relationships of a sample of individuals taken from a larger population
- Describes a probability distribution on ancestral genealogies (trees) given a population history, N(t)
- Can convert information from ancestral genealogies into information about population history and vice versa
- Is a model of ancestral genealogies, not sequences, and its simplest form assumes neutral evolution
- Can be thought of as a prior on the tree, in a Bayesian setting
Why the Coalescent Matters
The coalescent framework is crucial because it:
- Provides a null model for genetic variation under neutrality
- Enables inference of population parameters from genetic data
- Forms the basis for many modern population genetic analyses
- Connects microevolutionary processes to macroevolutionary patterns
2. Theoretical Population Genetics Foundation
The Wright-Fisher Model
Most of theoretical population genetics is based on the idealized Wright-Fisher model of population, which makes several simplifying assumptions:
- Constant population size N
- Discrete generations (no overlapping generations)
- Complete mixing (random mating)
- Neutral evolution (no selection)
- No mutation (in the basic model)
- No migration (closed population)
From Forward to Backward in Time
Traditional population genetics works forward in time, tracking allele frequencies through generations. The coalescent takes a different approach:
- Forward in time: Track all individuals and their descendants
- Backward in time: Track only the ancestors of the sample
This backward-time approach is computationally efficient because:
- We only track lineages that contribute to the present-day sample
- The number of lineages decreases as we go back in time
- We can ignore the vast majority of the population that left no descendants in our sample
3. Kingman's n-Coalescent
John Kingman's groundbreaking work in 1982 established the mathematical foundation for coalescent theory.
Consider tracing the ancestry of a sample of $k$ individuals from the present, back into the past. This process eventually coalesces to a single common ancestor (concestor) of the sample.
Kingman's n-coalescent describes the statistical properties of such an ancestry when $k$ is small compared to the total population size $N$.

The Coalescence of Two Lineages
Let's start with the simplest case: two randomly sampled individuals from a population of size $N$.
Two-Lineage Coalescence
- By perfect mixing, the probability they share a common ancestor in the previous generation is $1/N$
- The probability the common ancestor is $t$ generations back is:
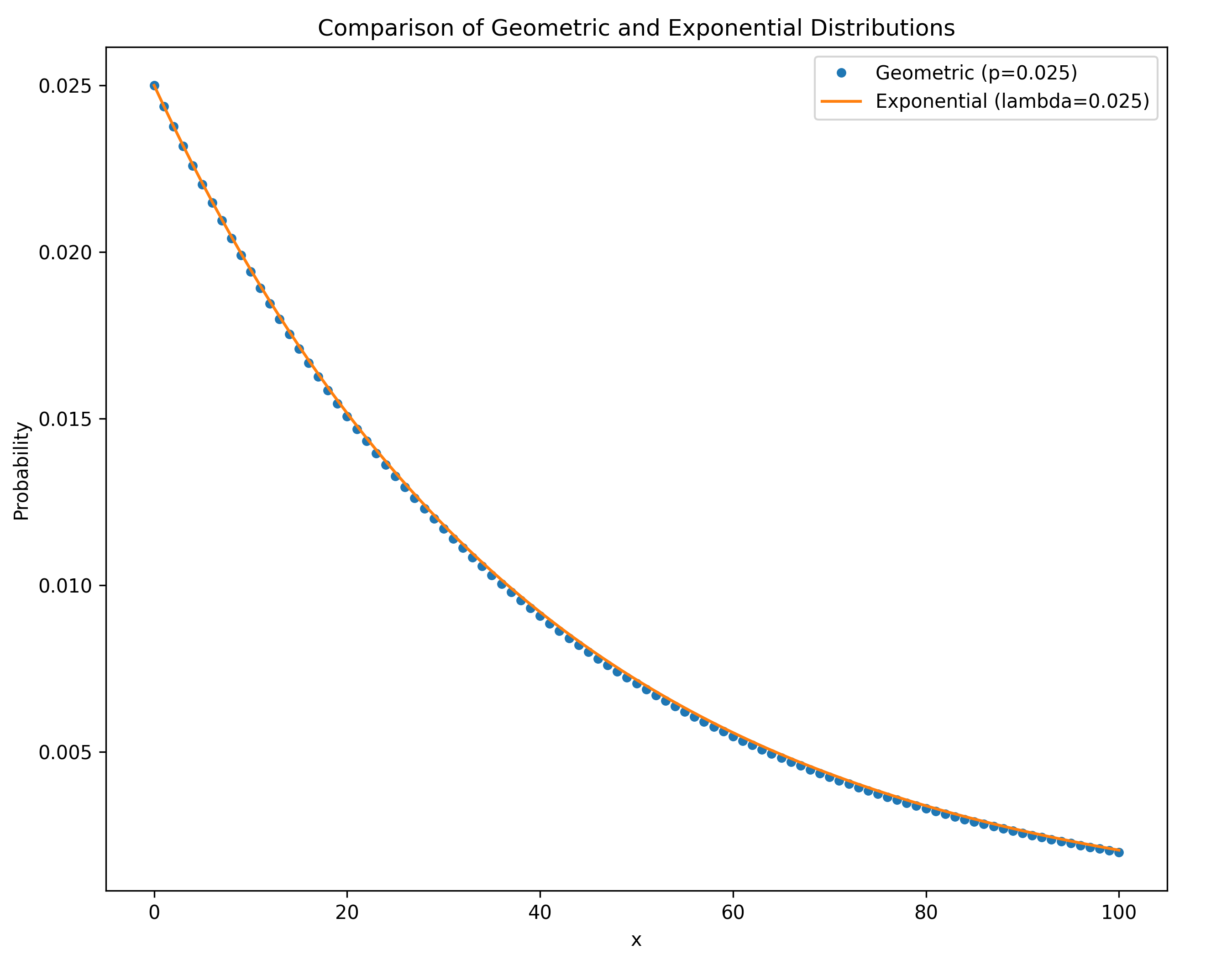
$$P(t) = \frac{1}{N}\left(1-\frac{1}{N}\right)^{t-1}$$
- This is a geometric distribution with success rate $\lambda = 1/N$
- Mean time to coalescence: $E[t] = N$ generations
- Variance: $\text{Var}[t] = N^2(1-1/N) \approx N^2$ for large $N$
The Coalescence of k Lineages
With $k$ lineages, the analysis becomes more complex but follows similar principles:
k-Lineage Coalescence
With $k$ lineages, there are $\binom{k}{2} = \frac{k(k-1)}{2}$ possible pairs that may coalesce.
- Coalescence rate: $\lambda_k = \binom{k}{2}/N$
- Mean time to first coalescence:
$$E[t_k] = \frac{N}{\binom{k}{2}} = \frac{2N}{k(k-1)}$$
- After coalescence, we have $k-1$ lineages remaining
The Complete Coalescent Process
The complete coalescent process for a sample of $n$ individuals involves successive coalescence events:
| Lineages | Coalescence Rate | Expected Time |
|---|---|---|
| $n → n-1$ | $\binom{n}{2}/N$ | $\frac{2N}{n(n-1)}$ |
| $n-1 → n-2$ | $\binom{n-1}{2}/N$ | $\frac{2N}{(n-1)(n-2)}$ |
| ... | ... | ... |
| $3 → 2$ | $3/N$ | $N/3$ |
| $2 → 1$ | $1/N$ | $N$ |
Total expected time to the most recent common ancestor (TMRCA):
4. The Coalescent as a Continuous-Time Process
From Discrete to Continuous Time
As the population size $N$ becomes large, the discrete-time Wright-Fisher model converges to a continuous-time process:
The Coalescent as a Diffusion Approximation
Kingman (1982) showed that as $N$ grows, the coalescent process converges to a continuous-time Markov chain.
Continuous-Time Coalescent
For $k$ lineages, the waiting time until the next coalescence follows an exponential distribution:
For a specific pair of lineages to coalesce:
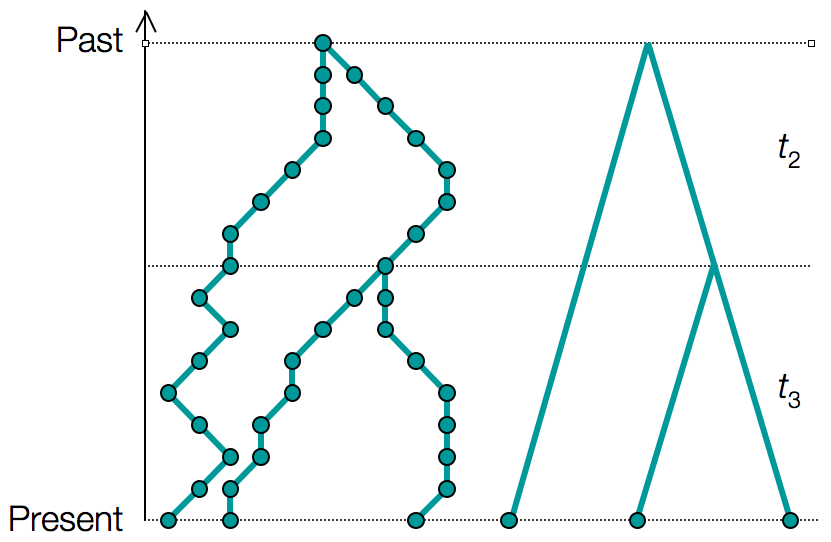
5. The Coalescent Density
Probability Density for a Genealogy
For a genealogy $g$ with $n$ samples and coalescent times $\mathbf{t} = \{t_2, t_3, \ldots, t_n\}$, we can write the probability density:
Coalescent Density Function
Where:
- $t_k$ is the time interval during which there are $k$ lineages
- The factor $1/N$ appears for each coalescence event (probability of specific pair coalescing)
- The exponential term accounts for the waiting time with $k$ lineages
Example: Three-Sample Genealogy
For $n = 3$ samples with coalescence times $t_3$ (3→2 lineages) and $t_2$ (2→1 lineages):
This shows how both topology and branch lengths contribute to the genealogy probability.
Properties of the Coalescent Density
- Factorization: The density factorizes over coalescence intervals
- Markov property: Given $k$ lineages, future coalescence is independent of past
- Exchangeability: All labeled topologies are equally likely under the standard coalescent
6. The Coalescent with Variable Population Size
Real populations rarely maintain constant size. The coalescent can be generalized to handle time-varying population sizes.
Variable Population Size Coalescent
Griffiths and Tavaré (1994) showed that for population size $N(t)$ varying with time:
Coalescent Density with Variable N(t)
The probability density for the first coalescence event at time $t$ given $n$ lineages:
Requirements:
- The coalescent intensity function $1/N(t)$ must be integrable
- $N(t) > 0$ for all $t$
Common Population Size Functions
Examples of N(t)
1. Exponential growth/decline:
2. Logistic growth:
3. Bottleneck:
Effects of Population Size Changes
Different demographic scenarios leave characteristic signatures in genealogies:
- Population expansion: Star-like trees with long terminal branches
- Population bottleneck: Rapid coalescence during the bottleneck
- Cyclic population size: Clustering of coalescence events
7. Applications and Extensions
The Coalescent as a Tree Prior
In Bayesian phylogenetics, the coalescent serves as a natural prior distribution on gene trees:
Where $\theta$ includes parameters like:
- Effective population size $N_e$
- Growth rate $r$
- Migration rates (for structured populations)
Extensions of the Basic Coalescent
- Structured Coalescent
- Accounts for population subdivision and migration
- Selection at Linked Sites
- Modifies coalescence rates due to hitchhiking effects
- Recombination
- Leads to the ancestral recombination graph (ARG)
- Multiple Merger Coalescents
- Allows more than two lineages to coalesce simultaneously
Practical Applications
The coalescent framework enables:
- Demographic inference: Estimating historical population sizes and growth rates
- Species delimitation: Distinguishing population structure from species boundaries
- Dating events: Estimating times of population splits or admixture
- Selection detection: Identifying deviations from neutral expectations
- Epidemiological inference: Tracking pathogen spread and transmission
Software Implementing Coalescent Models
- BEAST/BEAST2: Bayesian phylogenetics with coalescent priors
- MIGRATE: Estimation of migration rates and population sizes
- ms/msprime: Coalescent simulation programs
- LAMARC: Likelihood analysis with coalescent models
- IMa2: Isolation with migration models
8. Visualizing the Coalescent Process
Understanding the coalescent often benefits from visualization and simulation. The interactive demonstration in the lecture allows exploration of:
Key Parameters to Explore
- Sample size (n): How does increasing n affect the genealogy shape?
- Population size (N): How does N affect coalescence rate?
- Population dynamics: Effects of different N(t) functions:
- Sinusoidal (cyclic populations)
- Square wave (abrupt changes)
- Sawtooth (gradual change followed by crash)
Observable Patterns
Through simulation, we can observe:
- The randomness in coalescence times and tree shapes
- How bottlenecks accelerate coalescence
- The relationship between N(t) and the coalescent density
- Why most coalescence occurs near the present (for constant N)
Summary
The coalescent is a fundamental framework in population genetics that:
- Provides a null model: Describes genealogies under neutral evolution in idealized populations
- Works backward in time: More efficient than forward simulations for genealogical questions
- Links demography to genealogy: Population size changes leave signatures in gene trees
- Has elegant mathematics:
- Exponential waiting times between coalescence events
- Rate depends on number of lineages and population size
- Generalizes to variable population sizes
- Forms the basis for inference: Used as tree priors in Bayesian phylogenetics and for demographic inference
- Has many extensions: Can incorporate population structure, selection, recombination, and other biological realities
- Wakeley (2009) "Coalescent Theory: An Introduction" - Chapters 1-3
- Hein, Schierup & Wiuf (2005) "Gene Genealogies, Variation and Evolution" - Chapters 3-4
- Kingman (1982) "The coalescent" - Stochastic Processes and their Applications
- Griffiths & Tavaré (1994) "Sampling theory for neutral alleles" - Phil Trans R Soc B
Check Your Understanding
- Why is the coalescent more efficient than forward-time simulations?
- What is the expected TMRCA for a large sample from a population of size N?
- How does a population bottleneck affect the rate of coalescence?
- Why do all labeled topologies have equal probability under the standard coalescent?
- What information about population history can be inferred from genealogy shape?